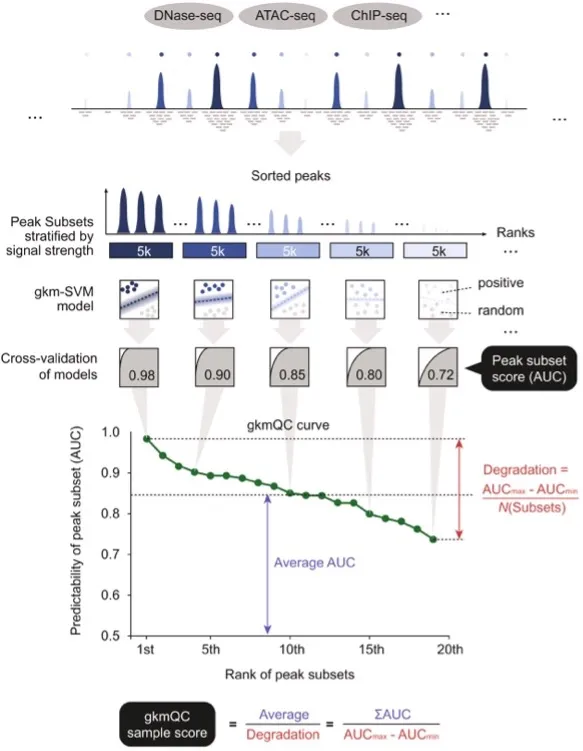
Chromatin accessibility assays are commonly used to identify regulatory elements in the genome that are associated with gene transcription. However, the quality of the data produced by these assays can vary significantly due to a range of biological and technical factors. To address this problem, we developed a machine learning method called gapped k-mer SVM quality check (gkmQC) to evaluate the quality of chromatin accessibility data using prediction accuracy as a metric. Using this method, we were able to identify "high-quality" samples that were more accurately aligned with functional regulatory elements and showed stronger associations with tissue-specific phenotypes. Additionally, gkmQC was able to optimize the peak-calling threshold for identifying additional regulatory elements, particularly in rare cell types. Overall, this study provides a useful tool for assessing the quality of chromatin accessibility data and identifying more reliable regulatory elements in the genome.