Research
Long-term Research Vision: Artificial Intelligence Applied to Quantitative Live Cell Imaging
The explosion of cell biology data generated by advances in microscopy, particularly live cell, super-resolution, and 3D imaging, as well as single-cell technologies, has revealed a high degree of heterogeneity that presents a major challenge for biomedical research. Artificial intelligence (AI) has made significant progress in the analysis of large, high-dimensional datasets and has demonstrated the ability to outperform humans in certain cases. However, current AI applications in cell biology have largely focused on static datasets such as single-cell RNA-seq and immunofluorescence images, rather than dynamic information from high-resolution live cell images.
To address this gap, we are developing an AI platform that can identify subtle or unknown dynamic phenotypes in live cell movies that are not readily discernible by the human eye. By utilizing this platform, we aim to unravel the phenotypic heterogeneity of cellular morphodynamics and motility in angiogenesis and cancer, and to facilitate precision cancer diagnosis and therapeutics.
AI Platform for Phenotyping Cellular Morphodynamics/Motility
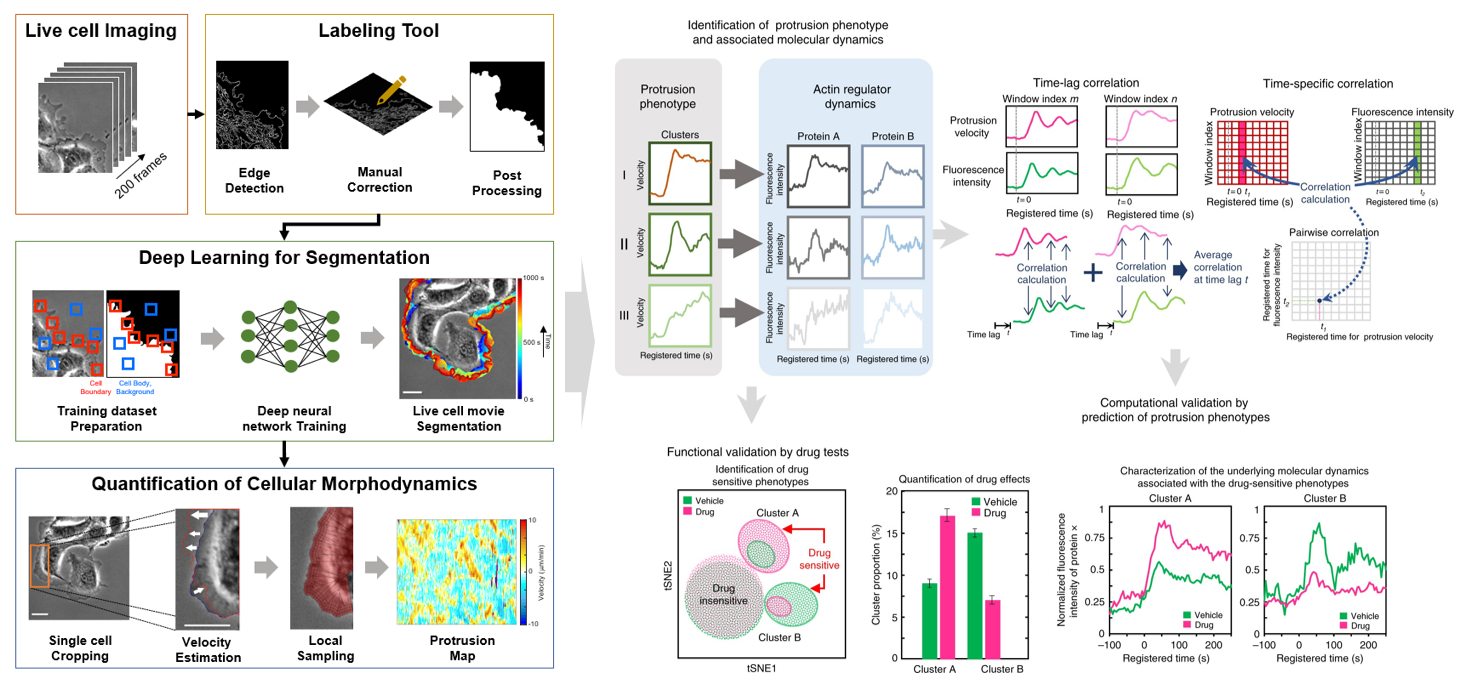
- Deep learning to deconvolve phenotypic heterogeneity of cell protrusion and retraction at the subcellular and single-cell levels
- High-througput live cell image acquistion and analysis by deep learning
- Deconvolution of heterogeneous effects of genetic/pharmacological perturbations
- Drug discovery based on cellular morphodynamics for angiogenic therapy
AI-based Breast Cancer Diagnosis
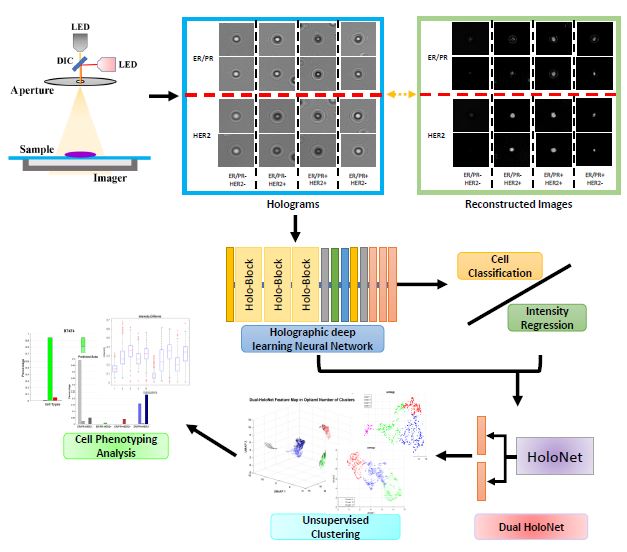
- Deep learning analysis of lensless inline holography images
- Dissecting the heterogeniety of breast cancer cells based on molecular markers
- Develop live-cell markers for breast cancer diagnosis
System Understanding of Cellular Morphodynamics Using AI
- Integrated analyses of the dynamics of cellular morphology, cytoskeleton (actin & microtubule), adhesions, mechanical force, and matrix degradation in endothelial or cancer cells
- Investigate how mechanical force affects the coordination of multiple cytoskeletal systems and morphodynamic phenotypes during endothelial/cancer cell migration
Collaborations
- Prof. Yongho Bae (Department of Pathology and Anatomical Sciences, University at Buffalo): Machine Learning Analysis of Spheroid Morphology
- Prof. Joe Italiano (Vascular Biology Program, Boston Children's Hospital/Harvard Medical School): Mordphodynamic Analysis of Megakaryocytes
- Prof. Hakho Lee (Center for Systems Biology, Massachusetts General Hospital/Harvard Medical School): Computational Imaging and Deep Learning
- Prof. Marsha Moses (Vascular Biology Program, Boston Children's Hospital/Harvard Medical School): Urine Biomarker Discovery
- Prof. Mauricio Santillana (Department of Physics, Northeastern University): Spatiotemporal Forecasting of COVID-19 Spread
- Prof. Ruby Wang (Division of Pulmonary Medicine, Boston Children's Hospital/Harvard Medical School): High-throughput Phenotyping of iPSC-derived Airway Epithelium
- Prof. Randy Watnick (Vascular Biology Program, Boston Children's Hospital/Harvard Medical School): Morphodynamics Analysis of Cancer Stem Cells
- Profs.Eun Hyun Ahn and Deok-Ho Kim (Department of Biomedical Engineering, Johns Hopkins University): Analysis of collective cancer invasion.
Funding
- Ongoing: NIH/NIGMS R35 (PI), NIH/NHLBI R01 (PI), NIH/NHLBI R01-subaward (Co-I), NIH/NCI R01-subaward (Co-I), BCH Start-up (PI)
- Completed: NIH/NIGMS R15 (PI), DoD Breast Cancer Research Program (PI), NSF (Co-PI)


