Serge Didenko Vasylechko, Simon K. Warfield, Sila Kurugol, Onur Afacan
Computational Radiology Laboratory
Boston Children’s Hospital and Harvard Medical School
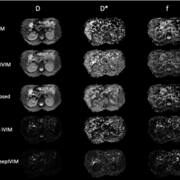
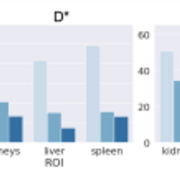
Deep learning methods have the potential to improve quantitative MRI methods. However, performance of deep learning methods is highly sensitive to the amount of available training data. In this work, we propose generating a substantial amount of 3D synthetic data, and demonstrate its application to myelin water fraction mapping. A parameter sampling model is designed within a naturally occurring range of multi-component T2 distributions to generate a large set of varying synthetic signals. This model is combined with a spatially varying sampling model that generates a multitude of spatial deformations and signal perturbations.